 This long interview may be difficult for some. In it Dr. Satterfield presents evidence which he believes suggests that XMRV will probably not work out. We’ve had this interview sitting in our laps for about 2 1/12 weeks during which we tried to get a response from the WPI . They, however, are too busy and when their Public Relations office was unable to give us a timeline for a response, we decided to publish it.
This long interview may be difficult for some. In it Dr. Satterfield presents evidence which he believes suggests that XMRV will probably not work out. We’ve had this interview sitting in our laps for about 2 1/12 weeks during which we tried to get a response from the WPI . They, however, are too busy and when their Public Relations office was unable to give us a timeline for a response, we decided to publish it.
Background – For the past 9 months or so a debate concerning the role PCR should play in the search for XMRV in chronic fatigue syndrome (ME/CFS) has arisen in the CFS community. The PCR results were the first results given in the Oct. 2009, Science paper and given PCR’s dominance in medical research field, it was not surprising that the research community tried to duplicate those results first (and then antibody results).
The senior author of the Science paper, Dr. Mikovits, however, has argued that culture, an older and more time consuming procedure that few labs use now, which does involve PCR, is a far better means of detecting XMRV and, indeed, the WPI’s now uses only culture and antibody tests to find XMRV in its ME/CFS patients. Thus far few attempts have been made to duplicate the WPI’s culture tests and none of them have cultured the sample for the time span Dr. Mikovits asserts is needed.
Dr. Satterfield https://www.codiagnostics.com/about/team.php comes from the PCR side. A summa cum laude graduate in Biomedical Engineering from Arizona State University he received his Ph.D in the same topic. Throughout his career he has been firmly ensconsed in the PCR arena and has developed numerous products to enhance PCR’s effectiveness. He is the founder, CEO and President of Cooperative Diagnostics , a medical diagnostic lab based in South Carolina.
Dr. Satterfield’s creation of new fast acting diagnostic probes with enhanced sensitivities which were able to withstand high temperatures apparently evoked interest from Homeland Security and he has worked with United States Army Medical Research Institute of Infectious Disease, Department of Homeland Security, National Biodefense Analysis and Countermeasures Center.
The Cooperative Diagnostics Lab was formed in January 2008 and currently provides real-time PCR tests for HIV, HCV, HBV, Dengue fever, Malaria, Chagas disease and Syphllis. It also provides a service to produce custom designed PCR tests in less than two months. https://www.codiagnostics.com/tech/custom.php. According to its website it utilizes statistical bioinformatics tools to deliver ‘novel molecular diagnostics” and boasts of several innovations including “Rapid Probes”, “Hot Sta-RT”, “Adaptivex Primers”, “Rapid Detex Primer’ and as well as proprietary statistical tools developed by Dr. Satterfield.
Cooperative Diagnostics and CFS – Cooperative Diagnostics has a substantial history with XMRV and ME/CFS. Using its rapid PCR test development technology it was the first lab to put a commercial test on the market. (This test was later withdrawn after the lab was repeatedly unable to find the pathogen in patient samples). Cooperative Diagnostics produced studies indicating it was unable to detect XMRV using antibody and PCR tests in people with autism in Oct 2010 and in CFS patients in Feb 2011.
This interview looks at a number of technical subjects and then closes with Dr. Satterfields personal experiences with ME/CFS.
CDC Study – the WPI gave the CDC positive samples and the CDC was unable to find XMRV in them. Doesn’t that prove that the CDC didn’t know how to find the virus? Even if XMRV was a contaminant or if mouse DNA was contaminating those samples, as some have suggested, why wasn’t the CDC able to pick that up? Doesn’t the fact that the CDC didn’t find anything mean that there was no mouse contamination?
Unfortunately, details on that analysis have not been released. It is difficult to say what exactly was done or was not done. Assuming that report is correct, it would imply that if contamination is involved it likely did not happen until some point after sample collection (ie during the DNA extraction or subsequent testing). Of course, the opposite could be true, that the CDC doesn’t know how to detect XMRV.
Really, that question is precisely what is under debate – is it incompetence on the part of scientists from around the world who cannot find XMRV, or contamination on the part of those who do find it? The idea that the CDC or others tested samples that were labeled as positive by the WPI certainly implies that things like geography or patient classification are not the reason why labs are getting different results. It all comes down to who is having false results.
PCR and XMRV
There’s a lot of talk about using a ‘wild-type’virus to devise a PCR test. What about that?
There have been a number of criticisms leveled against researchers using the XMRV VP62 strain for the purpose of designing a test. Clearly the test would need to detect the VP62 strain, but the design of the test should encompass much more to ensure that variant strains are not missed. Ideally, a test is designed to not just a single strain, VP62, but also to a number of different XMRV and MLV’s to help identify the regions of the virus that are truly common to the entire family of viruses.
 The interesting fact in all this is that the Lombardi et al test was the only one designed to look for just XMRV. There are some regions that are common to all MLV’s including XMRV and are what define an MLV. Lombardi et al actually avoided these regions to try to find a test that would only detect XMRV, presumably because the presence of MLV’s could be due to contamination.
The interesting fact in all this is that the Lombardi et al test was the only one designed to look for just XMRV. There are some regions that are common to all MLV’s including XMRV and are what define an MLV. Lombardi et al actually avoided these regions to try to find a test that would only detect XMRV, presumably because the presence of MLV’s could be due to contamination.
Since all the subsequent studies were more interested in maximizing the ability of the tests to find XMRV variants, they used the more stable, identifying regions of the MLV family for test design. This means that out of all the tests designed, the Lombardi et al test is the least likely to pick up variant strains.
Criticism has also been leveled at some researchers for not using “real” samples to validate their tests, which is a separate issue from that described above. The criticism is that instead of using samples identified as positive by Lombardi et al, they have used VP62 plasmid and/or XMRV infected 22Rv1 cells spiked into real samples. However, this is a catch 22.
If the objective of the study is to find out if Lombardi et al had a correct result (ie whether they had true positives or false positives), then how can their positive samples be used as the basis for the design of a new test? Doing that would assume that those positives had already been validated as true positives, which would defeat the entire point of the study!
 So, in order to establish the performance of any test, a “true positive,” which all researchers agree is positive, must be used. The VP62 plasmid and the XMRV infected 22Rv1 cell line are examples of positive controls that 100% of researchers can agree are positive.
So, in order to establish the performance of any test, a “true positive,” which all researchers agree is positive, must be used. The VP62 plasmid and the XMRV infected 22Rv1 cell line are examples of positive controls that 100% of researchers can agree are positive.
Another point is that in the establishment of detection limits (ie how sensitive a PCR test is), a spiked in control must be used. This is because it is important to know exactly how much has been added to each sample to calculate the detection limit.
Plasmids, such as the VP62 plasmid, are the ideal way to do this because it is very easy to calculate exactly how much you are adding to the sample. There really isn’t any other option for scientists in determining the detection limit.
Finally, a lot of people are probably wondering why someone doesn’t just take some positives from the Lombardi et al study and look for XMRV there. That should be a really easy way of deciding whether the Lombardi et al study was correct or not.
I think the problem there is one of trust. How do you decide who gets to design the experiments and whether the study has been done correctly or not? That is the importance of the Blood Working Group, which has taken input from all sides. It provides a fair, unbiased playing ground for Lombardi et al to show that what they found really holds up and to investigate differences in test performance by others.
There has been a lot of talk about doing a true replication study. You don’t seem to think a ‘true’ replication study with culture is necessary in order to determine if XMRV is present. Would you say that the PCR test in the original study has been replicated? When you say in the study that you used the same PCR tests as did Lombardi at all what are you saying? Are you saying you looked for the same gene sequences in both passes of the PCR?
That’s a very good question Cort. The main finding in the Lombardi et al study was the 67% of persons found to be XMRV positive by PCR. The reason all the other tests were included was for support of the main finding. All together Lombardi et al painted a pretty convincing picture. However, as in any structure, if the foundation is faulty, the whole argument collapses. Consequently, what scientists are interested in replicating is the key finding of PCR positives.
 In order to do a good study, a very specific question has to be asked. The question asked by scientists in the field is, “Can PCR of at least 100 to 250 ng PBMC DNA really detect XMRV in 67% of persons with CFS?” In fact, an even more basic question can be asked, “Can PCR of at least 100 to 250 ng of PBMC DNA detect XMRV in any CFS samples?” If it could, then proof of concept of XMRV infection could be established. Scientists could argue about the exact statistics of infection later on.
In order to do a good study, a very specific question has to be asked. The question asked by scientists in the field is, “Can PCR of at least 100 to 250 ng PBMC DNA really detect XMRV in 67% of persons with CFS?” In fact, an even more basic question can be asked, “Can PCR of at least 100 to 250 ng of PBMC DNA detect XMRV in any CFS samples?” If it could, then proof of concept of XMRV infection could be established. Scientists could argue about the exact statistics of infection later on.
There have now been quite a few papers looking for XMRV by PCR using at least 100 to 250 ng of PBMC DNA in persons with CFS and controls. So far in peer-reviewed literature, the results have been negative with one exception. Ironically, the one group that did not replicate the Lombardi et al conditions because they used less than 100 to 250 ng of PBMC DNA found MLV’s, but not XMRV, in an even larger number of persons with CFS.
So yes, the criteria of searching for XMRV in persons with CFS and controls by PCR using at least 100 to 250 ng of PBMC DNA has been replicated ad nauseum. And yes, what is meant by our paper and others that say they have used Lombardi’s test is that they have used the same primers, searching for the same gene sequence in XMRV.
You stated you used 3.3 – 8.8 times the DNA as in the original papers to look for the real-time PCR pol gene. What do you mean when you say you used more DNA? Does that mean you’re searching through more material or ‘blood’?
We used between 10 and 83 times as much DNA as the Lombardi et al and Lo et al studies in our pol test.However, since we used DNA from whole blood and compared our results principally to the Lombardi et al finding, we qualified that by stating that those large amounts of DNA contained between 3.3 and 8.3 times as much PBMC DNA as was used by Lombardi et al.
What we mean by more DNA is precisely what Dr. Mikovits said when she called for screening of larger “haystacks.” If you can find 67% positive by screening a drop of blood that only contains 100 to 250 ng of PBMC DNA, then what can you find screening 3.3 to 8.3 times that much? In the article you quoted earlier, she said that even detecting 67% positives, the amount of XMRV present was near the limits of detection of her PCR test, so we, and the authors in most of the other studies, created tests with better detection limits.
A recent article in BioTechniques.com called “Walking on Eggshells: PCR Assays for XRMV detection” reported that
“To increase the sensitivity of their nested PCR protocol, Mikovits turned to Max Pfost, a graduate student in her lab. Pfost researched PCR optimization and began modifying the protocol’s magnesium concentrations and annealing conditions, and choose primers for increased sensitivity rather than specificity.
Mikovits allegorizes from what she calls “the HIV days,” when it was first discovered that multiple low–copy number HIV strains existed. “If you are looking for a low–copy number family member, you really have to optimize the magnesium and base everything on the annealing temperatures.” Otherwise, she says, researchers using nested PCR would very likely overlook XMRV in their samples. “A lot of people are just not taking the time to optimize their PCR,” says Pfost”
What about this…are labs simply not taking the time to optimize their PCR as Pfost suggests?
In the late 1980’s, shortly after the invention of PCR, there were a lot of failed experiments due to a lack of understanding on how to optimize the tests. Since then, however, optimization has been a basic principle taught to undergraduates and anyone who works in a PCR lab. You simply cannot run reliable PCR tests without optimization.
I think most people would understand the logic that you can’t just pick up a book on the ABC’s of rocket design and then go tell the engineers at NASA that they’ve got it all wrong. Unfortunately, that is the very situation we have here with respect to the ABC’s of PCR diagnostics.
Professionally speaking, you don’t send your graduate student to read a book on something that you don’t know how to do, only to accuse all the professionals in the field of not understanding the most basic elements of the technique.
In the Biotechniques.com paper Dr Mikovits argued that culturing was necessary because the virus was so rare that you have to grow it out to find it many times.. She said “If you simply look for proviral DNA and look for very low–copy number virus, you have to make the cells divide”. Her graduate student who does the PCR said
“Some of these newer technologies are just not cutting it. You have to go back to culturing the virus…It’s hard to find the needle in the haystack if you’re not looking in a big enough haystack.”. Finally Dr. Mikovits said “ we’re talking about the detection limits of PCR” with XMRV.
PCR was what was used to find XMRV in 67% of samples in the Lombardi et al paper. Was this a real finding or not? If the authors of the Lombardi et al paper feel that this finding was made in error, then they should say so. That would save a lot of time and money on the part of scientists around the world. PCR is either sensitive enough to find XMRV as was done by Lombardi et al, or it is not and Lombardi et al were wrong.
ANTIBODIES
What about the antibody test? Antibody tests have always been purposed to be the ultimate test for XMRV and the WPI was able to find antibodies to XMRV and in about the same percentage as PCR positives. It seems like the likelihood of that happening by chance is kind of astronomical. Yet you had different antibody results than Lombardi et al. How can you explain the difference between the original antibody results and yours?
The antibody study in the Lombardi et al study was developed to support the PCR finding of 67% positives. The fact that Lombardi et al found that the samples were also antibody positive really seemed to prove that their work was correct. What was more, in 2009 when the study was published, there was evidence that XMRV had indeed integrated into clinical prostate cancer. All this data together was practically irrefutable.
Bring that forward to 2011 (where there are conflicting PCR results and data suggests the integrated XMRV was actually from contamination by XMRV infected cell culture) and the antibody testing is the last real leg to stand on.
No one disputes that antibody tests are the best method of detecting a chronic infection such as a retrovirus. Within a few weeks after a person is infected by a virus they develop antibodies to fight the viral infection. Those antibodies usually persist well after the infection has been cured and are found in much higher numbers than the viral DNA, making them easier to detect. Since retroviruses are not cured, antibodies to retroviruses are considered to be present for life, making them the most sensitive form of testing for retroviral infection.
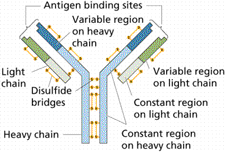 To make an antibody test, most labs use a fragment of a viral protein (fragments cost less money to produce and are easier to sell as tests to other labs). When that fragment comes in contact with the patient’s blood, if there are any antibodies present, they will stick to the protein. This is one of the oldest, most reliable forms of testing there is.
To make an antibody test, most labs use a fragment of a viral protein (fragments cost less money to produce and are easier to sell as tests to other labs). When that fragment comes in contact with the patient’s blood, if there are any antibodies present, they will stick to the protein. This is one of the oldest, most reliable forms of testing there is.
The problem with antibody testing is that sometimes the viral proteins used react with antibodies developed to other viruses. In fact, Lombardi et al relied on the principle of cross reactivity to take an MLV protein, hoping that it would be cross reactive with XMRV antibodies.
Given the cross reactive nature of antibody tests, the irony is that in order to make one of these super sensitive tests, PCR must first be used to identify samples that are truly positive. Since the PCR of the Lombardi et al study is under serious scrutiny for reliability, there are no positive samples that would qualify for designing an antibody test.
If it turns out that those samples do not have XMRV present other than from a contaminant, then no XMRV antibodies could have been formed. Any test designed to those samples runs the risk of being optimized to cross react with antibodies common in some other infection in people with CFS. In other words, it would be a test designed to have the maximum number of false positives in CFS patients.
The closest thing that we have to known positive samples is the blood from monkeys that were infected with XMRV. Since it is known which monkeys were infected with XMRV, and which were not, these can be used to make a research quality test.
The beauty of the antibody test created by the CDC is that they isolated live XMRV from cell culture and used each of the real XMRV proteins to search for antibodies. Since antibodies could theoretically be formed to any one of the proteins present in XMRV, using each of the proteins from actual XMRV has the maximum probability of finding XMRV antibodies if present. As far as sensitivity in antibody testing is concerned, it doesn’t get any better than that.
What’s more, the CDC antibody test is the only published antibody test used to evaluate CFS samples to have been validated on what are accepted by 100% of scientists as positive samples (ie the XMRV infected monkeys). As you know, we found no antibodies to XMRV in any of our CFS patients.
The WPI, though, has been working on an antibody test I believe and Dr. Mikovits reported that Dr. Bagni at the NCI has been working on a good antibody test as well. One report suggested that she may be validating the WPI’s results. What about those newer tests?
Again, it is important to use samples which are known to be positive to calibrate the test. Since the question of whether there are established XMRV positive samples from humans is still up for debate, calibration of a new antibody test to anything other than the known XMRV infected monkey samples could lead to false positive results.
CONTAMINATION
The WPI has never used any of the cell lines or reagents known to be contaminated with XMRV. They used separate rooms, separate buildings, separate hoods and cross-linked amplicons, tubes, and water supplies. They ran negative plasmas in every culture to prove that no steps were contaminated and they used mitochondrial DNA to test for contaminants. How could a contaminant sneak through all those precautions?
No one will deny that the Lombardi et al paper was done in as reasonable of a manner as can be expected for a first paper. Are there things that could have been done better in it? Sure, but there are in every paper. If all the other studies that had been done had found XMRV too, then the issue of contamination would have probably never come up. It’s just that since the adoption of PCR as a gold standard, every time there have been major discrepancies in PCR testing, it has been a result of contamination.
 Purely from a historical perspective, this looks like contamination. In the historical cases, the source of contamination was not immediately obvious either. Just like XMRV, those with the disease showed up positive more frequently than healthy controls in several labs. But in the end, it was all proven to be contamination.
Purely from a historical perspective, this looks like contamination. In the historical cases, the source of contamination was not immediately obvious either. Just like XMRV, those with the disease showed up positive more frequently than healthy controls in several labs. But in the end, it was all proven to be contamination.
How were those cases proven to be contamination? Well in at least one example, when the sequenced integration sites for the retrovirus came back, it turns out that instead of seeing human DNA on either side of the virus, there was rabbit DNA. That is why people like Dr. Coffin have been clamoring for proof that XMRV in persons with CFS has really integrated into the patients’ cells.
Unfortunately, the evidence that XMRV has integrated into the cells of patients with prostate cancer appears to be on shaky ground. Analysis of XMRV sequences shows they likely came from 22Rv1 cells. The 22Rv1 cells themselves have now been shown to have been contaminated. Also, at least a few of the sequenced integration cites from prostate cancer have been due to experimentally infected DU145 cell contamination.
In science, every paper is like a piece of a jigsaw puzzle. At first, what it’s going to look like is not clear. But as each piece is added, it makes a picture. You have a real good idea what it is going to look like even before the last few pieces have been added.
While the initial publications that came out were not real clear how they were going to affect the XMRV finding, the cumulative work has put together a pretty clear picture. In science, we usually say the data speaks for itself. Anyone looking at the picture that the data from the XMRV publications makes is going to have serious questions about contamination, even though the piece of the puzzle showing exactly where the contamination came from has not been found yet.
Multiple Avenues for Contamination Needed? – You make the argument that if you look at the situation from a kind of a statistical viewpoint that XMRV could work out but that it’s probably not going to. But it still seems difficult to explain contamination. If XMRV contaminated a batch of reagents then the next batch wouldn’t be contaminated – and they would know something was up. Or if XMRV is in the WPI’s lab – what about the NCI lab the UK study was done in that had never seen a mouse before? Those samples had never seen the WPI – yet they got results similar to those found at the WPI. Then there’s the testing at VIP Dx. Do we need different types of contamination for all these sites and how likely is that?
As stated previously, the discrepancies in PCR results between labs have happened before. Historically it has always been due to contamination. In the case of XMRV/MLV, there is already data showing a variety of sources of contamination affecting everything from PCR to sequencing.
While all the details in the puzzle have not been sorted out yet, there are some that have. Multiple labs have now reported that what they initially thought was XMRV/MLV was not. Each contamination event came from a different source. Some are as simple as primer-dimer based false positives in the PCR test. Some are contamination introduced from a common component used in PCR. Some contamination is detected by mouse mitochondrial tests. Others show a contaminant that isn’t obvious without testing for mouse genomic DNA, such as the IAP test.
But more intriguing than all these is the finding that both 22Rv1 cells and DU145 cells have likely contributed to contamination providing false positives in both the sequencing of XMRV and its integration sites. XMRV/MLV might be the first instance of a viral finding with such a high level of demonstrated contamination coming from what appear to be multiple sources.
So in answer to your question, yes there are multiple sources of contamination that can lead to XMRV/MLV false positives. And it is not only likely that there are different sources of contamination, but it is now proven and published in multiple peer-reviewed publications that there are different sources.
Controls – With all this controversy one would think the WPI must periodically be testing healthy controls to ensure that everything was kosher. If contamination was present the healthy controls would have the same positive rates as the people with CFS. Dr. Mikovits said ““The head of our Immunology Department congratulated me on being able to not contaminate the controls and only contaminate the patient samples in literally the thousands that we’ve done now,” How do you account for negative controls?
 I hate to sound like a broken record, but this has happened historically. What’s more, we don’t have access to the data that the WPI collects. We don’t know if the controls are being treated the same or differently. In the only blinded follow ups that I have seen the WPI participate in, one with the blood working group and another with the authors of one of the negative studies, they had false positives in the negative controls. Unfortunately both instances I am referring to only had one or two controls.
I hate to sound like a broken record, but this has happened historically. What’s more, we don’t have access to the data that the WPI collects. We don’t know if the controls are being treated the same or differently. In the only blinded follow ups that I have seen the WPI participate in, one with the blood working group and another with the authors of one of the negative studies, they had false positives in the negative controls. Unfortunately both instances I am referring to only had one or two controls.
We all look forward to seeing the data in the upcoming blood working group study. If the WPI can show consistent results (ie the same result in several blinded studies) in picking out people who have CFS from those who don’t, then that truly will be difficult to explain. Positive results from the blood working group study is probably the only way the WPI will be able to recapture the attention of the scientific community at this point.
22RV1 CELL LINE
There has been some talk about whether XMRV might have come from the 22RV1 cell line. Not long after XMRV was found Dr. Dusty Miller at the Fred Hutchinson Cancer Institute found, to his great surprised, that his prostate cancer cell line was growing XMRV. Then Dr. Hue found that the published sequences of XMRV were very similar to 22RV1. Could XMRV have come from this cell line and what does it mean if it does?
A lot of information on this subject has been published recently. Hue et al in a statistical analysis showed that the XMRV sequences that have been published are 95% likely to have originated from the 22Rv1 cell line.
That’s a pretty hard statement to digest. An easier way of looking at it might be thinking about identical twins. If you ran into someone you didn’t know who looks exactly like you and found out that their DNA is identical to your own, do you wonder at the odds that could have happened, or do you go question your parents about your newfound identical twin they never told you about?
 It’s the same odds and the same science that links XMRV to the 22Rv1 cells. Hue et al basically came out and said that there was greater DNA similarity between the XMRV sequences found in Lombardi et al and the prostate cancer studies than there was between the XMRV “identical twins” found in 22Rv1 cells.
It’s the same odds and the same science that links XMRV to the 22Rv1 cells. Hue et al basically came out and said that there was greater DNA similarity between the XMRV sequences found in Lombardi et al and the prostate cancer studies than there was between the XMRV “identical twins” found in 22Rv1 cells.
This then becomes something of a chicken or the egg question. Did the XMRV infection of 22Rv1 cells come from a clinical patient or were the positives from clinical patients really contamination from 22Rv1 cells? While the chicken and the egg question doesn’t seem to have an answer, the XMRV/22Rv1 cell question apparently does.
At the 2011 Conference on Retroviruses and Opportunistic Infections data was presented showing that when the 22Rv1 cells were originally isolated from the prostate cancer patient, they did not have XMRV. It was not until the cells had been passaged through nude mice various times that they acquired XMRV.
So in answer to which came first, it appears that at least the 22Rv1 cell line infection was in response to a contamination event and not from an actual prostate cancer infection. There is still some question as to where the 22Rv1 cell line may have gotten the XMRV from. More data was presented showing that XMRV could be a recombinant (ie a new virus created by two viruses that mix together) from two viruses present in mice.
While the statistics in the Hue et al study do not prove that the published clinical XMRV sequences came from 22Rv1 cell contamination, the 95% probability that is given is pretty high.
What about proving that XMRV is integrated into human cells. Why is that so important and how difficult is it?
Dr. Coffin and others have been publicly calling for this for a long time. Proving that the XMRV positives in CFS samples are integrated into human DNA would indicate that the positives are not mouse DNA contamination. It would make a huge difference.
You would still have researchers that are skeptical because it could still be contamination from the human 22Rv1 cell line or some other human cell line such as DU145 cells, but it would cause many researchers to start taking XMRV very seriously again.
Not every lab has the capability to identify the integration sites. For example, our lab doesn’t have the capability to do that, but other labs can do it in a couple of days. You need to have a reasonable quantity of virus in the sample in order to do it, but with the hundreds of positives the WPI has, surely at least one of the samples should have enough virus to check for integration into human cells.
COOPERATIVE DIAGNOSTICS
Cooperative Diagnostics XMRV Test – Cooperative Diagnostics came out with a test for XMRV pretty quickly. It used a very small quantity of blood – too small, Dr. Mikovits thought – to be able to pick up the virus. What about the low amount of blood used in the test?
Regarding the quantity of blood used in our commercial test we assumed that Lombardi et al had correct data; that PCR was capable of detecting XMRV in 100 to 250 ng of PBMC DNA. The blood spots we used hold 750 ng of DNA from whole blood, which contains 250 ng of PBMC DNA.
 We also thought this amount was incredibly small for detecting a virus given our experience with the blood viral levels of HIV and other viruses. However, we trusted the Lombardi et al data. As it turns out, we should have validated their finding before launching the test. We knew our test could pick out a single copy of XMRV in 250 ng of PBMC DNA contained in 750 ng of DNA from whole blood; what we didn’t know was if Lombardi et al was correct about XMRV being present in persons with CFS in only 100 to 250 ng of PBMC DNA.
We also thought this amount was incredibly small for detecting a virus given our experience with the blood viral levels of HIV and other viruses. However, we trusted the Lombardi et al data. As it turns out, we should have validated their finding before launching the test. We knew our test could pick out a single copy of XMRV in 250 ng of PBMC DNA contained in 750 ng of DNA from whole blood; what we didn’t know was if Lombardi et al was correct about XMRV being present in persons with CFS in only 100 to 250 ng of PBMC DNA.
Once the first peer-reviewed publications came out supporting what we were seeing in our lab, we realized that Lombardi et al was likely seeing contamination. So we pulled our test. We don’t want to be in the business of selling tests to people for a virus that doesn’t even exist in the human population.
However, we continued to search for XMRV in the blood of a number of volunteers using a variety of more sensitive testing methods. We wanted to make sure that if Lombardi et al were wrong about XMRV being present in 100 to 250 ng of PBMC DNA that we didn’t miss the virus being present at lower levels somewhere else.
Specifically, we used PCR to look for XMRV in 2,500 ng of DNA, up to a 25 times larger “haystack” than what was used by Lombardi et al and up to 83 times more than was used by Lo et al. Comparing apples to apples, it was up to 8.3 times more PBMC DNA than used by Lombardi et al.
In addition to those tests, we ran another 2 PCR tests and 2 RT-PCR tests (these look for RNA). Probably most compelling was the antibody test. In all known human retroviruses, antibodies appear a few weeks after infection and last for life. They are detectable even when viral levels drop below PCR detection limits. Of course we have already talked about why this antibody test was particularly relevant in response to one of your previous questions.
The details of this research and the results are now available in two peer-reviewed articles. One was on XMRV in autism and the other on XMRV in CFS.
How were you able to develop your tests so quickly? How could you find XMRV in such a small quantity of blood? How much did you use in your latest study?
Said simply, this is our profession. We are paid to understand PCR and to design PCR tests. We have invented a number of advancements in the field.
It’s an entirely different thing to go out and research what someone else did than it is to take what you have developed and apply it. The level of knowledge is different. That’s the difference between a Master’s degree and a Ph.D. In a Master’s, you learn what others did. In a Ph.D., you have to create new knowledge.
 My Ph.D. was obtained by developing systems of math that can screen literally millions of possible diagnostic designs to arrive at the best candidates. I applied that Ph.D. knowledge developing diagnostics for the Department of Homeland Security at various national security labs.
My Ph.D. was obtained by developing systems of math that can screen literally millions of possible diagnostic designs to arrive at the best candidates. I applied that Ph.D. knowledge developing diagnostics for the Department of Homeland Security at various national security labs.
Mostly they gave me what they considered to be impossible challenges fraught with false negatives and false positives. They wanted to see if my algorithms could solve what their entire department could not. When I would return two weeks later with results they could not obtain in years, they were speechless.
Eventually I started a company with that background as my base. Technology for me has always been a gift from God. It is not meant to be used to enhance your own self worth, but rather to lift the world. We have spent most of our time developing tests for applications overseas where we feel our technology is most needed. However, XMRV was different.
Why did Cooperative Diagnostics enter the field?
I might get a little personal here to answer your last question about why we entered this field. We all have friends in our lives. Some of those friends mean more to us than others. As we look back on our friendships, perhaps those that mean the most are with people who have sacrificed to make a difference in our lives.
I had one such friend who really helped me through a dark time in my life. I don’t think it is incorrect to say that she literally saved my life. If I am able to do any good in the world, it will be because of her.
After she helped nurse me back to health, she encouraged me to leave on a two year mission for Brazil. A few months after I left, she wrote me a letter saying that she and her roommates all got some real bad illness. One of the roommates recovered. But, she and her other roommate went on to develop something called “Chronic Fatigue Syndrome.”
I cannot tell you what it was like to be in Brazil and to receive letters from the person who in some ways I quite literally owe my life to, and to hear how this unheard of disease had literally crippled her. I had never met anyone who spent so many hours a day trying to help other people. Yet, there she was reduced to full time bed rest. Anytime she tried to do any activity, she was crippled even worse through the phenomena of “post-exertional malaise.”
 Over the next two years, I heard story after story of ignorant doctors and a government that was simply not educated on this disease. I have always wanted in my heart to do something for her, and not just for her but for the many people I have met since then whose lives have been torn apart by CFS.
Over the next two years, I heard story after story of ignorant doctors and a government that was simply not educated on this disease. I have always wanted in my heart to do something for her, and not just for her but for the many people I have met since then whose lives have been torn apart by CFS.
Unfortunately, I was never led in the direction of CFS research. What I was good at was applying math to molecular diagnostics (ie PCR). I went on to start a company based on PCR technology, but the yearning to do something for CFS has never left my heart.
When news came out that Lombardi et al had found a new retrovirus that was involved in CFS, it seemed too good to be true. Finally a cause to the disease had been implicated! Others had published on it in prostate cancer and it seemed there was no way the finding could be wrong. And what’s more, it was a retrovirus found in blood, our area of expertise!
Sometimes it seems like God places certain things in your path. This appeared to be one of those.
I know that there has been some criticism about the motives of various researchers who have found no association between XMRV and CFS. But I think with few exceptions, most of us were actually biased towards finding XMRV. It was not easy for any of these individuals, including myself, to publish against that finding.
Given all the emotion that was pent up in the finding by Lombardi et al, I cannot tell you how much I have struggled at a personal level with the data that XMRV is not associated with CFS. I did not want the data we and others found to be true. It is the reason we have continuously gone back and developed better tests for XMRV. It is the reason we partnered with the CDC. I kept thinking, maybe we missed something. But at some point, we all have to accept the data for what it is.
As much as I wanted XMRV to be the answer for CFS, I have to acknowledge that it is better for everyone if the truth comes out. Right now the data from numerous peer-reviewed publications indicates that XMRV is more than likely not associated with CFS. It hasn’t been disproven completely, but the chances are not good. We had to report what we found, because it was the right thing to do.
As sad as it is that XMRV does not appear to be panning out, I still think XMRV was a step forward for the CFS community. It has brought the disease to the attention of politicians in a way that has never been done before. I mean, even if XMRV is completely proven wrong, who will want to take responsibility for allowing persons with CFS to donate blood again? That potentially permanent ban on blood donations says something about the level of recognition that XMRV has garnered for persons with CFS.
It has also brought it to the attention of numerous scientists. I can’t help but think that at least a few are asking the question, “If XMRV isn’t the cause, then what is?” I believe that good will come out of all of this. We just have to focus on the positives.
One of those positives is that there are more scientists now than ever before that are engaged in fundamental research in CFS. That has to continue. The more data that is available, the more hypotheses can be generated. The more hypotheses that are generated, the more likely that some will start to have a positive impact on discovering the cause of CFS, leading to therapies for symptom relief and eventually the cure. We have to keep pressing forward.
………..
